ORILINX
A genome language model to map DNA replication origins.
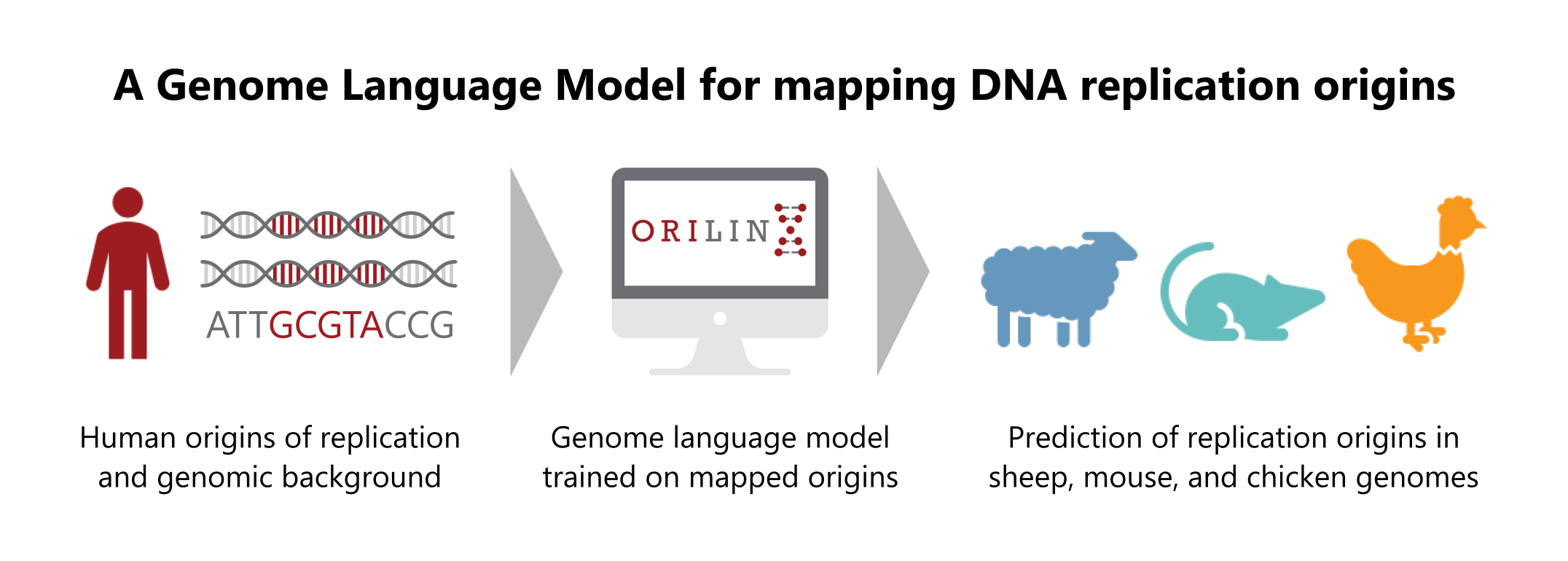
ORILINX is a genome language model for predicting DNA replication origins and initiation landscapes directly from primary DNA sequence. It addresses a long-standing question in human DNA replication: how initiation sites are specified in the absence of a defining DNA motif.
ORILINX is based on the DNABERT-2 genome language model, fine-tuned on experimentally mapped human replication origins from SNS-seq. By learning extended sequence context beyond simple compositional features, the model identifies both strong and weaker origins across the genome and captures sequence signatures associated with origin competence.
When applied genome-wide, ORILINX predictions closely align with independent estimates of origin efficiency inferred from replication timing and mathematical modelling, despite the model having no access to timing data. Although trained on human data, the model generalises to other vertebrates, accurately predicting origins in mouse, sheep, and chicken genomes.
Together, these results indicate that intrinsic DNA sequence encodes meaningful information about replication initiation and that genome language models provide a scalable way to extract this information. Check out our preprint, and try ORILINX.